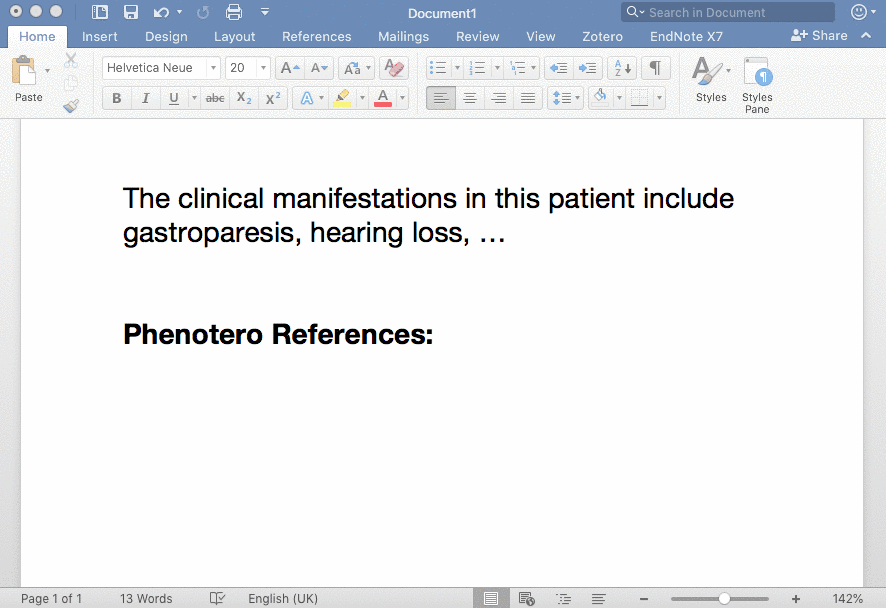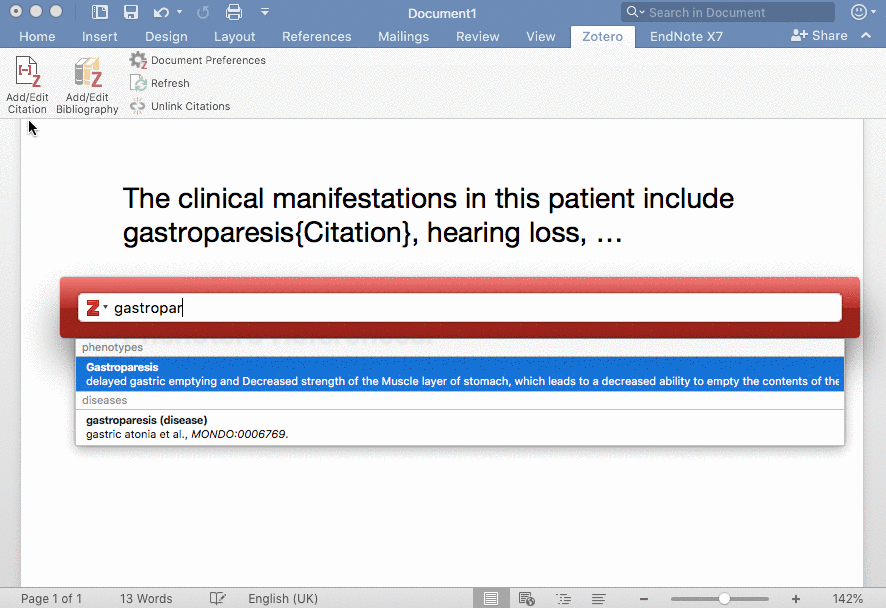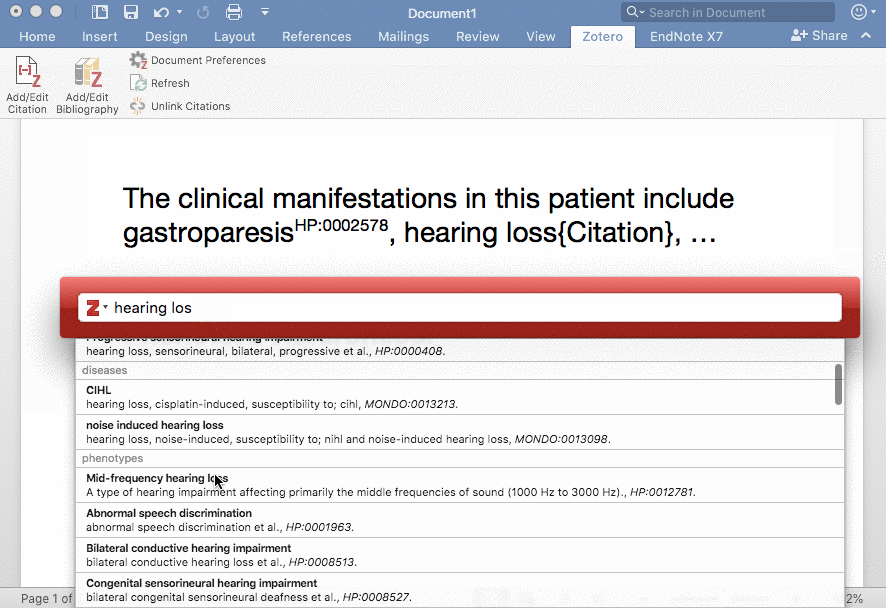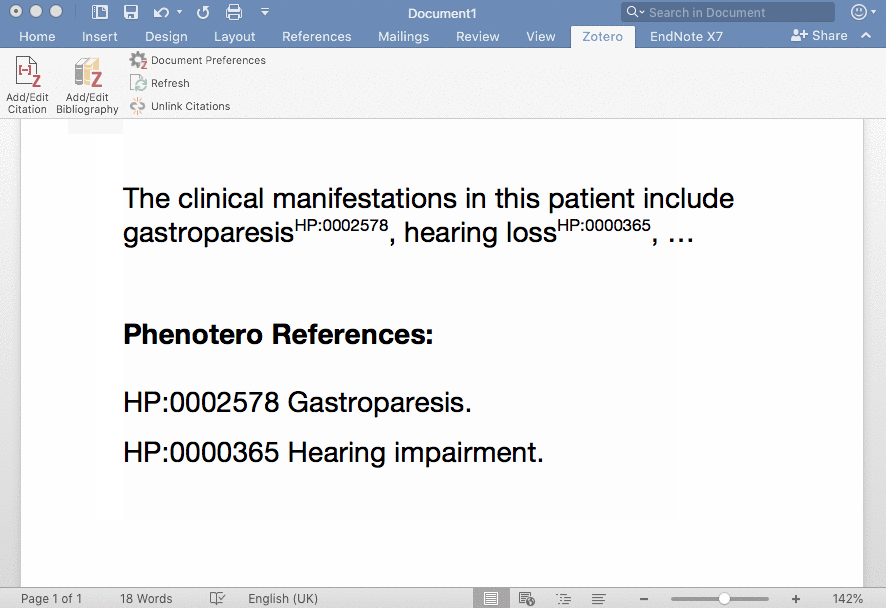
Phenotero is a simple tool for referencing phenotypes and diseases in written text and allows clinicians and clinical researchers to stay within word processing software. This enables a novel workflow for direct in-text annotations of ontology classes (such as HPO terms), which we call annotate as you write.
Zotero
Phenotero is based on the awesome, free, and open-source citation software Zotero and thus builds on existing features such as a word-processor plugin, standardised and extendible citation styles, a search functionality, and active community support.
Why Phenotero ?
Phenotero provides a quick and easy solution for referencing ontology-based phenotypes and diseases by direct in-text annotations of classes of ontologies such as the HPO or MONDO. It is an extension of the Zotero citation management open source software, which runs on all common operating systems and is easily integrated into multiple word processors, including LibreOffice, Microsoft Word and Google Docs. Users are presented with a minimal amount of computational complexity and can readily stay within their usual working environment.
Data & Documentation
Please find links to all data and to the documentation in the upper right corner of this web site.
The Creators
Phenotero is developed by the Translational Genomics Team and the Phenomics Team in Berlin. Specifically these are Daniela Hombach, Jana Marie Schwarz, Dominik Seelow, Markus Schuelke, Ellen Knierim, and Sebastian Köhler.
Publication
Further detailed information about Phenotero is available in our, which is available at https://onlinelibrary.wiley.com/doi/10.1111/cge.13471. We are happy about any kind of feedback on the manuscript as well as on the usage of Phenotero.
Related projects
If you have a professional informatics infrastructure, you might consider using PhenoTips. For analysing you text retrospectively, you might want to have a look at the Monarch annotator. (Note that for the latter your text is sent through the internet)
Feedback
Contact us at sebastian.koehler@charite.de.